 B2.1: Mechanism fo RNA Unwinding catalyzed by DEAD-Box Helicase
B2.1: Mechanism fo RNA Unwinding catalyzed by DEAD-Box Helicase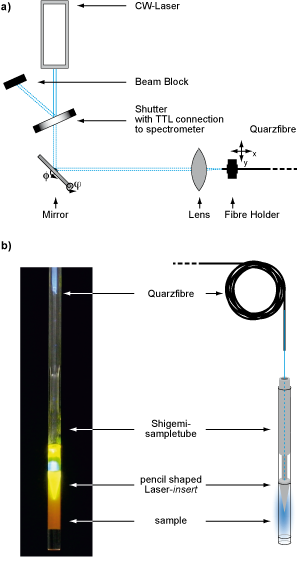
This PhD student will perform structural and mechanistic studies of a DEAD-Box helicase from in E. coli. In order to characterize the RNA remodeling mechanism of RNA helicases with high spatial and temporal resolution, static solution state and time-resolved NMR spectroscopy as tool for detection will be used. Additionally, to delineate events in the unwinding reaction on different timescales UV-VIS spectroscopy will be used in conjunction to NMR spectroscopy. This will be performed in collaboration with PhD project A3.3 of the Wachtveitl group.
After having established a protocol for the cloning and purification of the RNA helicase the PhD student will characterize the structure and dynamics of the helicase using high-resolution NMR spectroscopy in conjunction with differently caged ATP and RNA molecules.
The RNA-protein association reaction will be investigated by NMR spectroscopy, whereby the triggers of the process will be differentially caged ATP molecules. In a next step, the influence of the helicase on a RNA target-structure and the mechanisms of RNA unwinding will be delineated. These experiments that focus on the spatial resolution of the kinetics of the RNA unfolding reaction, will rely on an RNA target that contains a photo-removable road-block. He/She will receive the caged RNA from PhD project A2.3. The road-block allows association of protein with the RNA but hinders propagation of the unwinding reaction. From the halted RNA-protein complex the propagation of the unfolding reaction is initiated by photolysis. The read-out will then be made by real-time NMR and UV-VIS spectroscopy. The spatial resolution of the read-out experiment with both methods is achieved by the application of special labeling schemes.
The combination of the results of all the planned experiments will give new and deep insights in the mechanism of how ATP-dependent proteins shape the folding landscape of RNAs.
Figure: Laser Setup for in-situ illumination of NMR-samples a) laser setup outside the magnet, b) modified NMR sample tube
Suggested Literature:
- "RNA refolding studied by light-coupled NMR spectroscopy", H. Schwalbe, B. Fürtig, Methods Mol Biol. 2014, 1086, 309-319.
- "Transient RNA-protein interactions in RNA folding", M. Doetsch, R. Schroeder, B. Fürtig, FEBS J. 2011, 278, 1634-1642.
 B2.2: NMR of chaperone-assisted RNA refolding under light control
B2.2: NMR of chaperone-assisted RNA refolding under light control
In the first period of the CLIC-Program Heidi Zetzsche performed structural and mechanistic studies of the DEAD-Box helicase from in E. coli. After having established a protocol for the cloning and purification of the RNA helicase she characterized the dynamics of the helicase using high-resolution NMR spectroscopy in conjunction with caged ATP molecules. The technical emphasis was put on time-resolved NMR spectroscopy methods for characterization of the RNA remodeling mechanism. The helicase mediated RNA refolding reaction was thereby triggered by caged ATP molecules. Additionally, to delineate events in the unwinding reaction she collaborated with Dinh du Tran (A3.3) who performed UV-VIS spectroscopy analysis using fluorescently labelled target RNA molecules.
In the next term the focus will be shifted towards RNA chaperones, molecules that promote RNA refolding reaction but without relying on ATP as energy source. Again, the NMR spectroscopic methods developed during the first term will be applied and extended. Instead of triggering the energy source of the reaction, the RNA target molecule itself will be caged to generate a non-equilibrium state, which relaxation to its equilibrium will be followed both in the presence and absence of chaperone molecules. The synthesis of the caged RNAs will again be performed by PhD students of A1 from the Heckel group. Events occurring on fast timescales unresolvable by time-resolved NMR will again investigated in collaboration with the Wachtveitl group using their advanced UV-VIS spectroscopy methods. The combination of the results of all the planned experiments will this time give new and deep insights in the mechanism of how ATP-independent proteins shape the folding landscape of RNAs.
